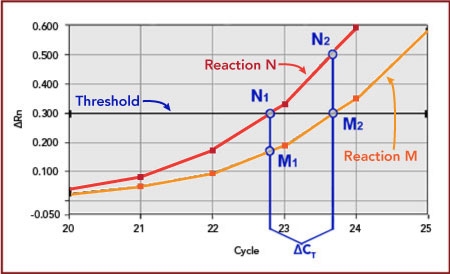
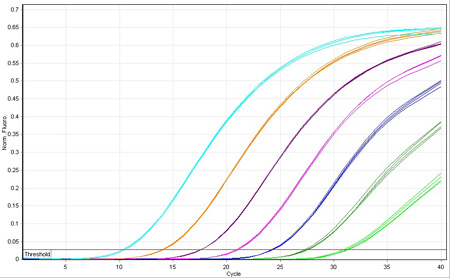
Since its invention, PCR has revolutionized the manipulation of gene sequences. With the introduction of quantitative PCR (qPCR) came the ability to measure the expression level of those genes with a precision and sensitivity not afforded previously.
Biosearch supplies the fluorescence-quenched probes and primers that that are the critical component to gene expression analysis. DNA replication triggers fluorescence from the probe so that the kinetics of the amplification can be correlated with initial copy number. Probes also provide additional sequence specificity to ensure that the monitored amplification is truly the gene of interest.
Following is a brief overview of the qPCR mechanism. We outline several different methods of quantification applied to gene expression analysis, but equally applicable toward other uses of copy number determination, for example, loss-of-allele analysis and assaying for viral load.
For more comprehensive and complete step-by-step guidance on working with custom oligonucleotides including how to handle these in the laboratory, optimisation and validation of assays, and batch control, please read our Guidance for optimisation and validation of custom oligonucleotides.