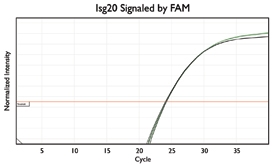
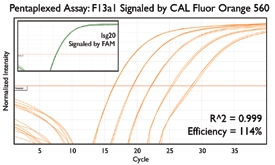
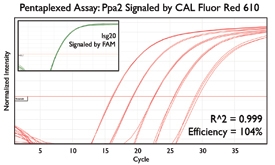
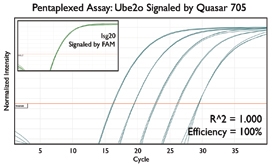
A variety of purposes are served by combining several assays together into one. Here are a few key examples from the literature.
Internal Positive Control
An internal positive control is a secondary assay that amplifies with every experiment, to confirm that genetic material is properly extracted and that cDNA is efficiently generated. Representing either an endogenous target such as a native gene or an exogenous template spiked into the sample, this control can be assayed in separate wells on the PCR plate but provides even more assurance if combined into the same wells for detecting your gene of interest. Only by multiplexing together with your other assays can you have complete confidence in every single reaction on your plate.
Development of an Internal Positive Control for Rapid Diagnosis of Avian Influenza Virus Infections by Real-Time Reverse Transcription-PCR with Lyophilized Reagents. Das, A.; Spackman, E.; Senne, D.; Pedersen, J.;Suarez, D.L. J Clin Microbiol. 2006; 44(9): 3065–3073.
Replicate Assays
Since the read-out from qPCR assays can occasionally mislead, some researchers detect the same gene with several distinct assays. This redundancy boosts confidence in quantification since several factors produce contradictory results: degradation of the RNA transcript, differing efficiencies during reverse transcription, and nonspecific amplification from off-target regions. Reagent handling is then streamlined by combining together these replicate assays into a single reaction, making more efficient use of pipetting steps, PCR reagents, and sample material.
Quantification of mRNA using real-time RT-PCR. Nolan, T.; Hands, R.E.; Bustin, S.A. Nat Protoc. 2006;1(3):1559-82.
Distinguishing Strains
Related organisms may share many similar sequences making them difficult to distinguish them from their nearest neighbors. For example, strains of microbes harboring certain plasmids demonstrate a virulence absent from relatives that are otherwise identical. Researchers amplify multiple signatures to properly identify these strains and diagnose their threat to health. If this molecular diagnostic becomes a routine test then multiplexing can facilitate commercialization so that fewer reactions are required to assign each result.
Multiplexed detection of anthrax-related toxin genes. Moser, M.J,; Christensen, D.R.; Norwood, D.; Prudent, J.R. J Mol Diagn. 2006 Feb;8(1):89-96.