Looking for oligos? Visit our new oligo and Stellaris dedicated platform at
oligos.biosearchtech.com
SNPviewer
General Information
SNPviewer is a tool that enables genotyping data to be viewed as a cluster plot (see below), but that does not include data analysis or reporting functionality. Genotyping data from a service project run at LGC Biosearch Technologies is typically sent to the customer as a SNPviewer file (in addition to a text file). Opening genotyping service project results in SNPviewer will allow you to view the genotyping clusters plate by plate. Genotyping calls cannot be changed within SNPviewer; if you need to be able to edit the calls then you will require our KlusterCaller software.
Resources
Your genotyping data may be visualised by downloading the SNPviewer PC tool. This will let you graphically view the clusters that group the allele calls. This software will present your results on a treeview by SNP and master plate. The plates may be clicked on to view the data. Calls can not be changed in SNPviewer. If you need to change any calling then you need KlusterCaller.
Minimum PC specification
SNPviewer 64-bits is suitable for computers that are running on Windows 10 64-bits.
Installation instructions
You will need administration rights on your PC to install SNPviewer.
- Click here to download - SNPviewer
- Right click the installer ‘SNPviewer.setup.27582.exe’ and select ‘Run as administrator’. Please note: you should first uninstall any existing SNPviewer-version from your machine. This can be done from ‘Control Panel’ > ‘Programs and Features’. Once you have done this, start the installation as an administrator.
- Work through the installation wizard to install SNPviewer on your machine.
How to use
To download our SNPviewer software (free-of-charge) please click here.
This SNPviewer help page covers the following topics:
- Getting started – opening SNPviewer and importing a .csv data file to view
- Exploring the genotyping data within SNPviewer –view a genotyping cluster plot, customise the menu tree, and search for an assay or plate ID within the project.
- Interpreting the genotyping calls – a guide to the colour codes used on the genotyping cluster plot
- View a project summary – viewing a summary of the project being viewed in SNPviewer including percentage call rate and total number of data points
- Export of data – a guide to customising genotyping data for export to a .csv file
- Export of cluster plot – exporting a cluster plot image from a genotyping project viewed in SNPviewer
Getting started
To open SNPviewer, click on the icon ![]() . Alternatively, select ‘SNPviewer’ and then ‘SNPviewer’ executable from the Start menu of Windows.
. Alternatively, select ‘SNPviewer’ and then ‘SNPviewer’ executable from the Start menu of Windows.
Once SNPviewer opens, you will be prompted to open a genotyping results file. Navigate to the data file (.csv format) that you wish to view, and click ‘OK’.
The data will be imported into SNPviewer.
Exploring the genotyping data within SNPviewer
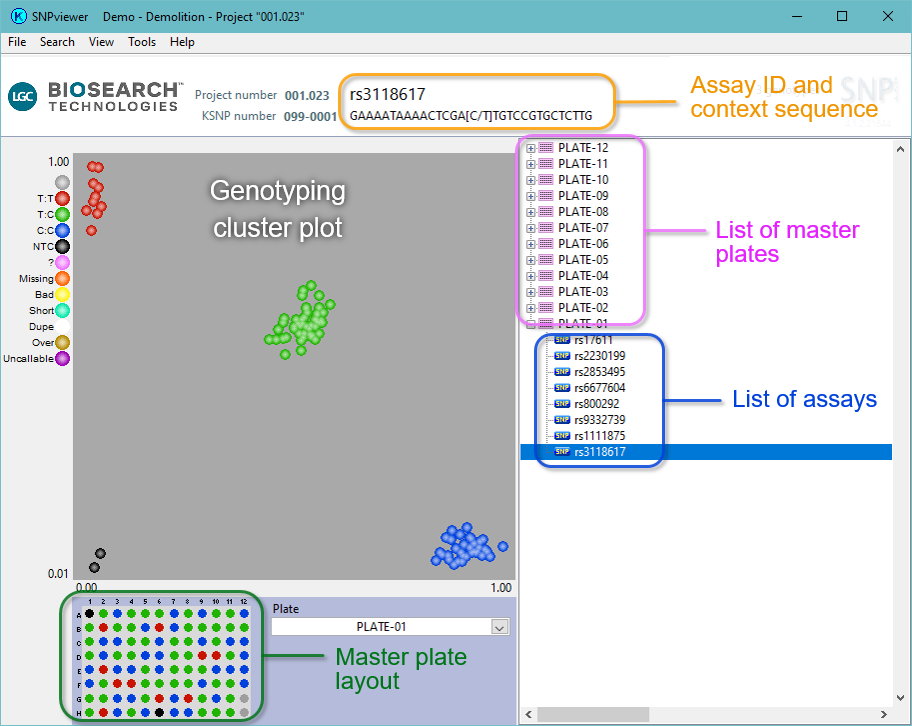
Once your data set has opened within SNPviewer, you will see a similar window to that shown in Figure 1. The DNA master plates and assays used in your project will be displayed in the menu tree on the right hand side of the screen.

Figure 1. SNPviewer window. In the tree on the right hand side, each assay and sample master plate is listed. The cluster plot is displayed by clicking on the relevant plate in the menu tree. Blue data points are homozygous for the allele reported by FAM, green data points are heterozygous and red data points are homozygous for the allele reported by HEX. The black data points represent the no template controls (NTC). The DNA sample plate layout is shown below the cluster plot.
To view the data for a particular assay or DNA plate, navigate to the relevant location within the menu tree and click on the plate / assay to display the genotyping cluster plot.
You can change the order within which the data tree is displayed, Click on ‘View’ in the top menu bar and select ‘View SNP or Plate list options’.
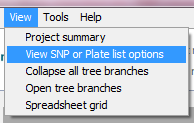
A dialogue box will open with the four available options for viewing data.
Select your preferred option and click ‘OK’.