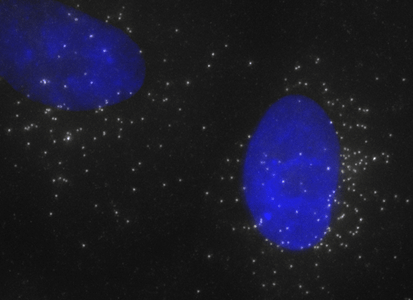
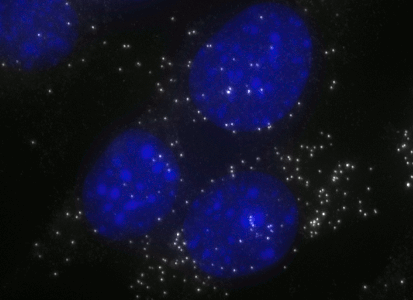
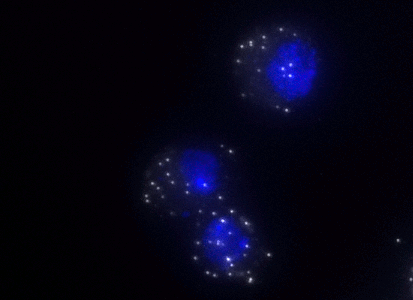
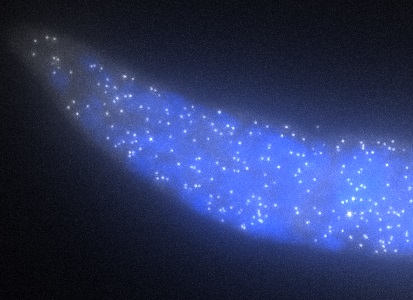
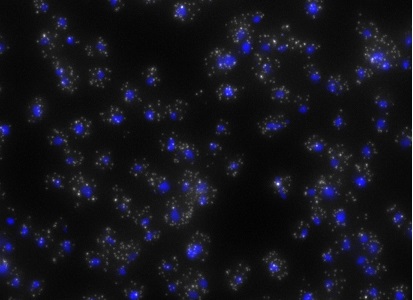
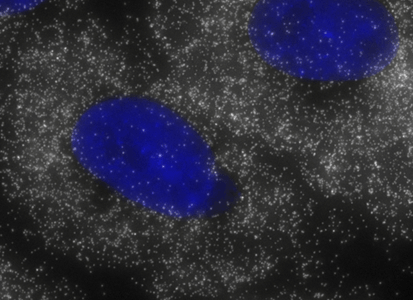
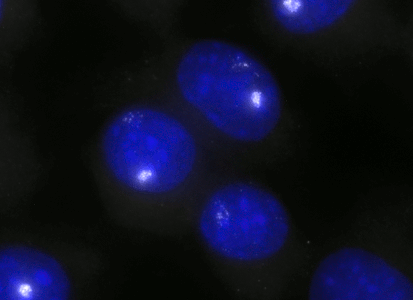
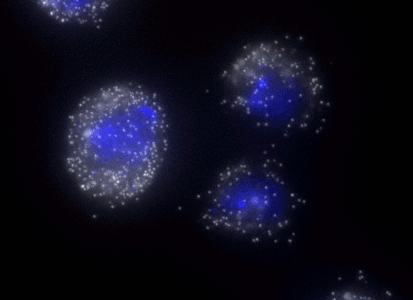
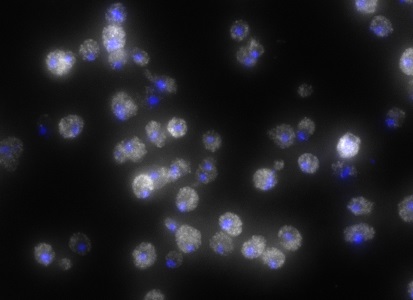
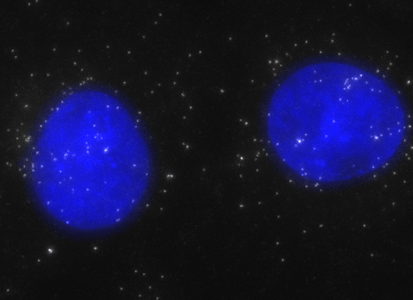
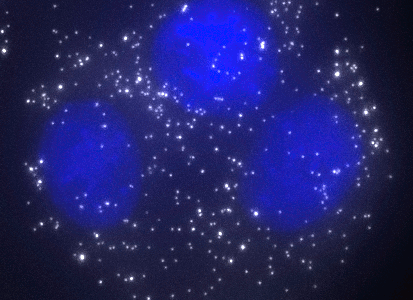
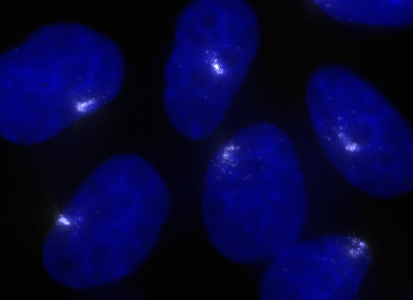
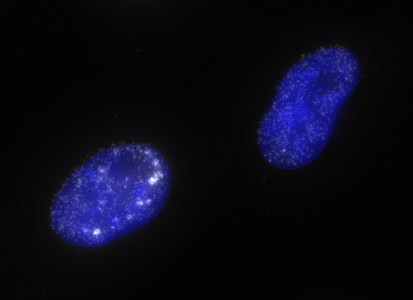
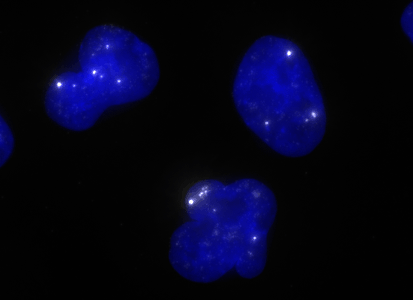
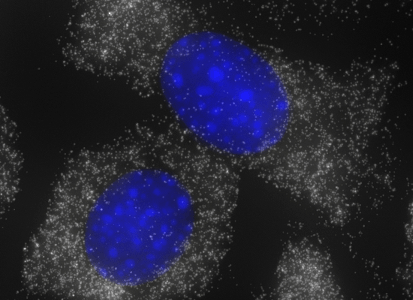
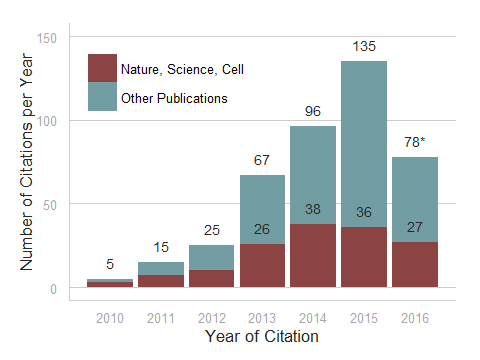
ShipReady control probe sets for Stellaris™ RNA FISH significantly improve the ease of use for the Stellaris technology by providing convenient, ready-to-ship positive controls against reference messenger RNAs and long non-coding RNAs (mRNAs and lncRNAs). Both qualitative (localization) and quantitative (transcript counting) controls are available in the most commonly used model organisms including Human, Mouse, C. elegans, Drosophila, and Yeast. Stellaris ShipReady Probe Sets recognize RNAs from previously validated common control genes, are ready to ship, and the delivered amount is 1 nmol of pooled oligos (sufficient for 40-80 hybridisations).
The sequence information of ShipReady Stellaris RNA FISH Probe Sets is proprietary to LGC Biosearch Technologies, and we do not disclose this information.